MOTIVATION
As we face the challenge of establishing a sustainable society and mitigating the effects of climate change on our ecosystems, it is imperative that we develop a much deeper understanding of how a diverse array of organisms perceives, responds and acclimates to climate-change associated stresses (1). In this regard, plants represent ideal model systems for expanding the frontiers of our knowledge. Despite a sedentary habit, plants traverse vast stretches of our planet and have evolved adaptive strategies to survive across every ecosystem. While general mechanisms of environmental response have been studied, important climate-change associated stresses such as drought and salinity still remain mysterious in how they are perceived and what adaptive mechanisms exist to ensure survival (2). The Dinneny lab is tackling these timely and broadly relevant questions through a multiscale approach that spans cell biology, comparative genomics and physiology, and synthetic biology. Our lab’s research aims to identify fundamental mechanisms used by plants to survive environmental stress, and to develop methods that allow us to model and tune such mechanisms through synthetic engineered organisms (3). Knowledge gained here will inform our general understanding of how cells sense physical and chemical changes in their environments, how the evolution of genomic sequences create novel physiological states through the rewiring of gene regulatory networks (GRNs), and how synthetic biology and machine learning can be used to explore structure-function relationships in organisms through the development of synthetic morphologies and signaling pathways.
RESEARCH STRATEGY
My lab is guided by three central tenets: (1) Water is the most limiting resource for life on earth, thus, the biology of how organisms deal with water availability represents one of the most significant processes we can hope to understand. (2) Organisms have independently evolved strategies to survive water-limiting conditions, thus the most broadly relevant physiological principles will be uncovered through a comparative approach. (3) Machine learning and synthetic biology approaches are needed to rigorously test structure-function relationships learned through our molecular-focused approaches. Together, these tenets have helped me to establish a dynamic lab that is both broad in expertise yet laser-focused on solving biological mysteries at the heart of what allows plants to be such masters of environmental acclimation.
CENTRAL QUESTIONS
How do plants know where water is in their environments? How do plants optimize their bodies and physiologies to thrive with a limited supply of water or a high concentration of salinity? How might we use this knowledge to discover and apply design rules for plant-water relations that can improve agriculture and sustain biodiversity? The questions asked and the methodological innovations needed to answer these questions are universal to life sciences with plants arguably representing some of the best organisms to focus on because of the integral role the environment plays in determining their physiologies.
PROJECT AREA 1: The cell wall—the missing link for moisture perception in plants?
For many organisms and cell types, the extracellular matrix is the direct interface with the environment (4, 5). In plants, this is especially so as all cell types are encased in a complex meshwork of polysaccharides, proteins and associated metabolites and ions. This matrix dynamically changes in composition in response to cellular cues associated with growth and differentiation but can also be directly perturbed by the environmental milieu outside of the cell (6, 7). While studies have identified control points for cell wall biosynthesis that are modulated by water-associated stress (4, 7, 8), whether the wall itself contributes to the perception of these environmental cues is unclear (9). In plant cells, the plasma membrane is physically appressed to the wall due to turgor pressure in the cell. Under hyperosmotic stress, turgor will subside and the membrane partially dissociates from the wall. It has been hypothesized that the change in wall-membrane association may be directly sensed by the cell as a means of detecting water availability (4, 10), however, this has not been directly tested. Understanding the mechanistic basis for communication between the wall and the cell will likely elucidate important principles driving a broad range of plant-environmental interactions, and more generally, will provide insight into the diverse ways that cells utilize mechanical and chemical cues originating from the extracellular matrix to regulate their behavior.
Crop relevance of cell-wall mediated moisture signaling: The cell wall is likely also involved in mediating the response of plants to water availability in crop plants. Previous research in my lab had identified a novel response of roots to water that we term hydropatterning (2, 15, 16). In soil, roots are exposed to substantial environmental heterogeneity with pockets of water and air associated with soil particles. We discovered that plant roots sense this heterogeneity at the micron scale and activate branching (lateral roots) along those root surfaces directly contacting water, while surfaces that contact air are inhibited from forming branches. Currently the mechanistic basis for how roots detect these differences in water availability are not known but our work using computational modeling of tissue hydraulics has indicated that external differences in water availability can be propagated to internal tissues but only in growing regions of the root tip (17). Furthermore, micromanipulation experiments have confirmed that the growth zone is the only region of the root competent to respond to moisture cues.
In the major crop maize, we have surveyed a diverse panel of inbreds and found substantial variation in hydropatterning that is well correlated with in-field root system architecture, which suggests that this trait is field relevant. We used these data to perform a Genome-Wide Association Study (GWAS) and identified the loci associated with cell wall signaling that we are currently characterizing to elucidate how proteins in the wall directly sense water status, while research in maize will allow us to understand how variation in hydropatterning affects the drought tolerance of plants in the field.
PROJECT AREA 2: Using biodiversity to understand how root systems enable adaptation to extreme environments.
Roots are the primary organs in plants that acquire and transport water to the above-ground shoot and are relatively simple in structure, consisting of concentric layers of tissue surrounding a central vascular cylinder (20). The cell layer immediately external to the vasculature is termed the endodermis, which accumulates hydrophobic polymers in its wall that block the passive diffusion of solutes, such as sodium, into the root through the extracellular spaces (21). While textbooks provide clearly delimited physiological roles for each tissue layer of the root, little is known regarding the diversification of such functions across species. Comparative anatomical studies of roots are rare and molecular genetic studies comparing gene functions between species have only recently been attempted (22, 23). Characterization of the anatomical diversity that exists in root systems is needed, but these efforts are best when paired with knowledge of the phylogenetic relationships of species and the soil and climatic conditions these species come from. Diversity in root functions also arise from variation in the growth rate, gravity response, and branching rate of roots. These processes are highly responsive to environmental and endogenous cues, which leads to the dynamic architecture of the root system (24). Structure-function relationships between specific root architectures and environmental conditions have been postulated but rarely tested rigorously (25).
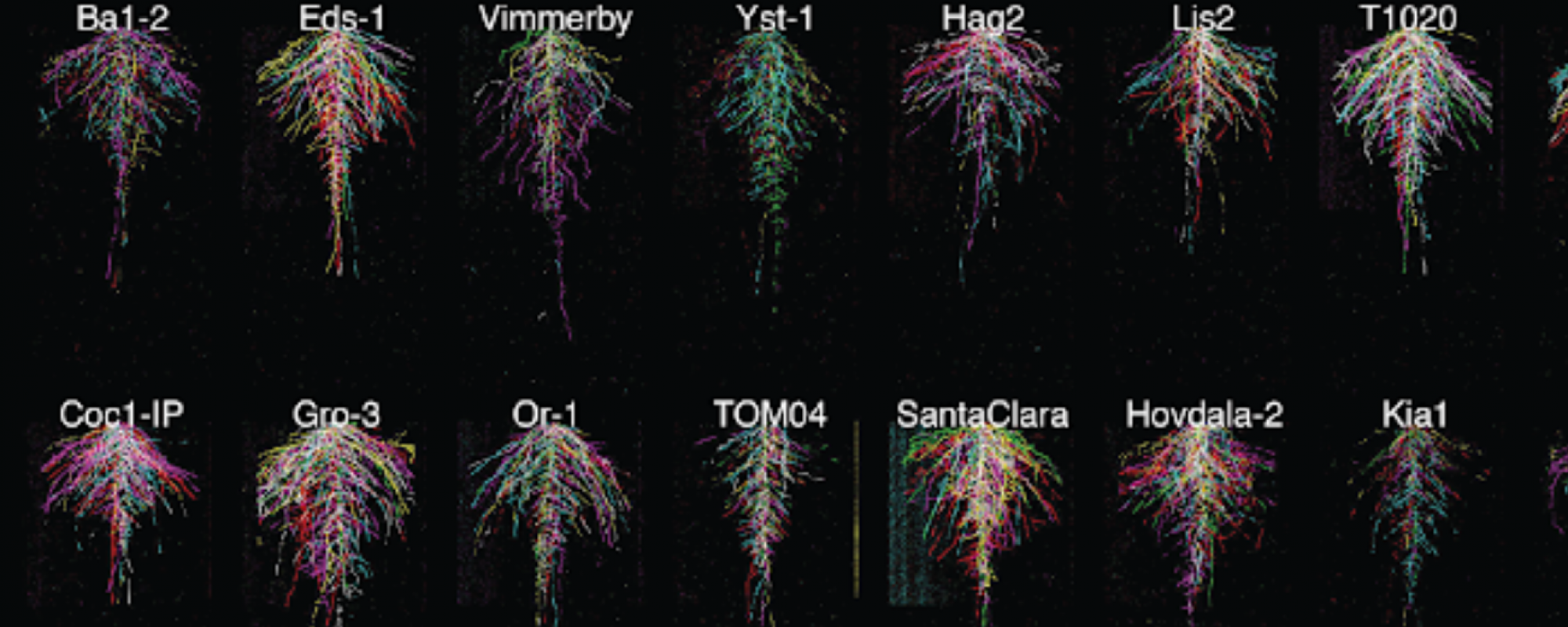
Establishing the Brassicaceae family as a tool for comparative physiology: To determine whether the genetic pathways that regulate the response to water are conserved across species and how the functions of roots in sensing and acquiring water are diversified to meet specific environmental challenges, we have established a phylogenetically informed collection of species in the Brassicaceae family amenable to molecular-genetic and genomic analysis (26). Relative to other plant families such as Solanaceae (tomato, tobacco) and Poaceae (maize, rice) families, Brassicaceae species have the advantage that most plants are annuals, transformable using the rapid floral dip method, and most importantly, closely related to Arabidopsis, which is by far the best system for molecular studies of gene function (27). Our Brassicaceae sub-collection comprises 11 species with sequenced genomes including oilseed crops: Camelina sativa (28), Thlaspi arvense (pennycress) (29), Brassica napus (rape seed) (30); molecular-genetic models: Cardamine hirsuta (31), Arabidopsis thaliana (32); extremophyte crop relatives: Schrenkiella parvula (33), Eutrema salsugineum (34, 35), Arabidopsis halleri (36), Arabis alpina (37); and phylogenetically informative species: Sisymbrium irio (38), Capsella rubella (shepherd’s purse) (39). We have established efficient transformation vectors and protocols for most species and have successfully performed CRISPR/Cas9 editing, which allows us to learn how gene functions are diversified.
Diversity in root anatomy, architecture and stress response in the Brassicaceae: We are currently establishing an anatomical atlas of the root tissues in our Brassicaceae sub-collection and using computer vision software to digitally encode this atlas to allow for the assessment of 3D tissue organization and computationally predicted physiological properties. Assays examining the response of species to the drought and salinity stress hormone abscisic acid (ABA) have revealed dramatic rewiring of this response across species and suggest that “stressful” environments may trigger growth promotion in some species adapted to such environments (40). This work is complemented with scRNAseq analysis of cell-type functional diversity (collaboration with John Schiefelbein, Univ. Michigan and Maheshi Dassanayake, LSU) that is revealing how changes in the number of tissue layers leads to differential partitioning of cellular functions in species.
A phylogenetically resolved Gene Regulatory Network (GRN) for stress response: Currently we are using this Brassicaceae sub-collection to establish how the GRN for drought/salinity is rewired across evolutionary time to generate physiological responses tuned to stressful environments. GRNs are constructed using DAP-seq, which is an in vitro method for establishing the genome-scale landscape of a transcription factor (TF) (41, 42). While DAP-seq does not work for every class of TF, we and others have found that the bZIP-class is highly amenable. Being able to do such assays in vitro allows GRNs to be constructed for non-model species where traditional ChIP-seq methods are technically challenging. Our initial work comparing 4 species revealed how changes in the GRN for the stress hormone ABA led to the differential control of cell elongation in some species, which accelerated root growth under salinity stress (40). We are expanding the GRN to include all 11 species and 6 additional DAP-seq-compatible TFs that cooperatively regulate stress-responsive gene expression. These data will allow us to track how the gain or loss of network connections over evolution led to the divergent stress response programs we observe today across the Brassicaceae phylogeny. Such knowledge will inform our understanding of how transitions between stress sensitive and resilient physiological states are driven by the rewiring of signaling networks as well as changes associated with crop domestication.
PROJECT AREA 3: Using machine learning and synthetic biology to understand structure-function relationships in roots
Just as computational modeling of development facilitates discovery by formalizing the quantitative relationships between molecular and physical systems in organisms (43), synthetic biology presents physiologists with the opportunity to discover the design rules for organ systems with a rigor difficult to achieve in the past (3). For example, while the role of root system architecture in determining the efficiency of water and nutrient capture is supported by physiological experiments and computational modeling (44), proof of these relationships often relies upon comparisons between existing crop varieties or wild species (45), which differ in more than just their root architectures. The ability to regulate gene expression with predictable accuracy using synthetic gene circuits promises to enable a more rigorous assessment of these relationships by the rational design of anatomical features in organisms and the experimental exploration of the function of these synthetic systems. Such approaches will be accelerated by machine learning (ML) which can be used to inform the design-build-test-learn cycle by facilitating the learning of patterns in biological systems (46), and the synthesis of promoters with defined regulatory activity through generative ML algorithms.
ML-based prediction and design of non-coding sequence function: An important goal of our work in the Brassicaceae family is to predict the physiological properties and stress resilience of an organism based on genomic sequence (47, 48). Achieving this ambitious target will require the data sets described in PROJECT AREA 2 as training data for AI-ML-based algorithms and the design of an experimental-basis for validation. Future projects will use these algorithms to evaluate the adaptive value of standing genetic variation in species under different climate change scenarios (49). Furthermore, generative ML algorithms are planned that compose de novo promoters with predictable regulatory activity under stress (46). Such approaches will inform our understanding of how non-coding genetic variation in a species contributes to the capacity to adapt to stress, and will enable the creation of synthetic promoters as inputs for gene circuits that regulate the architecture of roots.
Designing synthetic morphologies to test structure-function relationships in plants: In our recent work, we established a toolkit and methodological pipeline to design, build, and test synthetic gene circuits in plants to drive predictable patterns of gene expression and quantitatively tune the branching of roots (50). While important, this study represents just the beginning of our long-term goal to use synthetic gene circuits to generate morphological diversity for exploring structure-function relationships in plants. Current work is focused on adapting this approach to tune the branching rate of roots in grass species such as Setaria and Sorghum, which represent important models and crops for food and bioenergy production (51). Our synthetic tool kit will need to be reassessed in grass species, which is being done in collaboration with Jennifer Brophy (Stanford). Furthermore, we are now testing protocols that dramatically improve plant transformation efficiency in grasses, which will facilitate the large-scale testing of synthetic gene circuit architectures (52). Establishing plants that quantitatively differ in root architecture will allow us to test how branching rate determines access to water, but also whether we can detect a fitness cost associated with the metabolic burden of a larger root system (53, 54). Integration of logic gates to incorporate environmental regulation of root architecture will enable the engineering of root systems that reprogram development in response to new environmental stimuli. These studies will leverage the development of our GLO-Roots phenotyping platform that utilizes luminescence-based reporters to track the spatiotemporal growth patterns of root systems in soil-like conditions (55, 56). Robotics-based automation of watering and environmental conditions together with the ability to automatically image up to 96 plants per day will allow us to test realistic patterns of simulated precipitation and drought, which is impossible with traditional gel-based systems, and at a throughput impossible with more expensive X-ray microCT-based systems (57).
REFERENCES
- H. Hirt, S. Al-Babili, M. Almeida-Trapp, M. Antoine, M. Aranda, D. Bartels, M. Bennett, I. Blilou, D. Boer, A. Boulouis, C. Bowler, S. Brunel-Muguet, F. Chardon, J. Colcombet, V. Colot, A. Daszkowska-Golec, J. R. Dinneny, B. Field, K. Froehlich, C. H. Gardener, A. Gojon, E. Gomès, E. M. G. Álvarez, C. Gutierrez, M. Havaux, S. Hayes, E. Heard, M. Hodges, A. K. Alghamdi, L. Laplaze, K. J. Lauersen, N. Leonhard, X. Johnson, J. Jones, H. Kollist, S. Kopriva, A. Krapp, M. L.-P. Masson, M. F. McCabe, L. Merendino, A. Molina, J. L. Moreno Ramirez, B. Müller-Röber, M. Nicolas, I. Nir, I. O. Orduna, J. Pardo-Tomás, J.-P. Reichheld, P. L. R. Egea, H. Rouached, M. M. Saad, P. Schlögelhofer, K. A. Singh, I. De Smet, C. Stanschewski, A. Stra, M. Tester, C. Walshe, A. P. M. Weber, D. Weigel, P. Wigge, M. Wrzaczek, B. Wulff, I. M. Young, PlantACT! – how to tackle the climate crisis. Trends Plant Sci. 0 (2023), doi:10.1016/j.tplants.2023.01.005.
- J. R. Dinneny, Developmental Responses to Water and Salinity in Root Systems. Annu. Rev. Cell Dev. Biol. (2019), doi:10.1146/annurev-cellbio-100617-062949.
- J. A. N. Brophy, T. LaRue, J. R. Dinneny, Understanding and engineering plant form. Semin. Cell Dev. Biol. (2017), doi:10.1016/j.semcdb.2017.08.051.
- Y. Rui, J. R. Dinneny, A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. (2019), doi:10.1111/nph.16166.
- C. M. Nelson, M. J. Bissell, Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 22, 287–309 (2006).
- W. Feng, D. Kita, A. Peaucelle, H. N. Cartwright, V. Doan, Q. Duan, M.-C. Liu, J. Maman, L. Steinhorst, I. Schmitz-Thom, R. Yvon, J. Kudla, H.-M. Wu, A. Y. Cheung, J. R. Dinneny, The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca2+Signaling. Curr. Biol. (2018), doi:10.1016/j.cub.2018.01.023.
- L. Colin, F. Ruhnow, J.-K. Zhu, C. Zhao, Y. Zhao, S. Persson, The cell biology of primary cell walls during salt stress. Plant Cell. 35, 201–217 (2023).
- R. Gutierrez, J. J. Lindeboom, A. R. Paredez, A. M. C. Emons, D. W. Ehrhardt, Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806 (2009).
- I. Lang, D. A. Barton, R. L. Overall, Membrane-wall attachments in plasmolysed plant cells. Protoplasma. 224, 231–243 (2004).
- G. B. Monshausen, S. Gilroy, Feeling green: mechanosensing in plants. Trends Cell Biol. 19, 228–235 (2009).
- D. J. Cosgrove, Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 (2005).
- C. T. Anderson, J. J. Kieber, Dynamic Construction, Perception, and Remodeling of Plant Cell Walls. Annu. Rev. Plant Biol. 71, 39–69 (2020).
- T. C. Branon, J. A. Bosch, A. D. Sanchez, N. D. Udeshi, T. Svinkina, S. A. Carr, J. L. Feldman, N. Perrimon, A. Y. Ting, Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
- A. Mair, S.-L. Xu, T. C. Branon, A. Y. Ting, D. C. Bergmann, Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. Elife. 8 (2019), doi:10.7554/eLife.47864.
- Y. Bao, P. Aggarwal, N. E. Robbins 2nd, C. J. Sturrock, M. C. Thompson, H. Q. Tan, C. Tham, L. Duan, P. L. Rodriguez, T. Vernoux, S. J. Mooney, M. J. Bennett, J. R. Dinneny, Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. U. S. A. 111, 9319–9324 (2014).
- N. E. Robbins, J. R. Dinneny, The divining root: moisture-driven responses of roots at the micro-and macro-scale. J. Exp. Bot. (2015) (available at https://academic.oup.com/jxb/article-abstract/66/8/2145/496250).
- N. E. Robbins 2nd, J. R. Dinneny, Growth is required for perception of water availability to pattern root branches in plants. Proc. Natl. Acad. Sci. U. S. A. (2018), doi:10.1073/pnas.1710709115.
- H. Shi, Y. Kim, Y. Guo, B. Stevenson, J.-K. Zhu, The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 15, 19–32 (2003).
- G. J. Seifert, The FLA4-FEI Pathway: A Unique and Mysterious Signaling Module Related to Cell Wall Structure and Stress Signaling. Genes . 12 (2021), doi:10.3390/genes12020145.
- B. Scheres Benfey P. Dolan L., “Root Development” in The Arabidopsis Book, C. R. Somerville Meyerowitz E. M., Ed. (American Society of Plant Biologists, Rockville, MD, 2002), vol. doi: 10.11, pp. 1–18.
- N. Geldner, The endodermis. Annu. Rev. Plant Biol. 64, 531–558 (2013).
- C. Ortiz-Ramírez, B. Guillotin, X. Xu, R. Rahni, S. Zhang, Z. Yan, P. Coqueiro Dias Araujo, E. Demesa-Arevalo, L. Lee, J. Van Eck, T. R. Gingeras, D. Jackson, K. L. Gallagher, K. D. Birnbaum, Ground tissue circuitry regulates organ complexity in maize and Setaria. Science. 374, 1247–1252 (2021).
- K. Kajala, M. Gouran, L. Shaar-Moshe, G. A. Mason, J. Rodriguez-Medina, D. Kawa, G. Pauluzzi, M. Reynoso, A. Canto-Pastor, C. Manzano, V. Lau, M. A. S. Artur, D. A. West, S. B. Gray, A. T. Borowsky, B. P. Moore, A. I. Yao, K. W. Morimoto, M. Bajic, E. Formentin, N. A. Nirmal, A. Rodriguez, A. Pasha, R. B. Deal, D. J. Kliebenstein, T. R. Hvidsten, N. J. Provart, N. R. Sinha, D. E. Runcie, J. Bailey-Serres, S. M. Brady, Innovation, conservation, and repurposing of gene function in root cell type development. Cell. 184, 3333–3348.e19 (2021).
- R. Rellán-Álvarez, G. Lobet, J. R. Dinneny, Environmental Control of Root System Biology. Annu. Rev. Plant Biol. 67, 619–642 (2016).
- W. Feng, H. Lindner, N. E. Robbins 2nd, J. R. Dinneny, Growing Out of Stress: The Role of Cell- and Organ-Scale Growth Control in Plant Water-Stress Responses. Plant Cell. 28, 1769–1782 (2016).
- P. E. Verslues, J. Bailey-Serres, C. Brodersen, T. N. Buckley, L. Conti, A. Christmann, J. R. Dinneny, E. Grill, S. Hayes, R. W. Heckman, P.-K. Hsu, T. E. Juenger, P. Mas, T. Munnik, H. Nelissen, L. Sack, J. I. Schroeder, C. Testerink, S. D. Tyerman, T. Umezawa, P. A. Wigge, Burning questions for a warming and changing world: 15 unknowns in plant abiotic stress. Plant Cell (2022), doi:10.1093/plcell/koac263.
- N. J. Provart, J. Alonso, S. M. Assmann, D. Bergmann, S. M. Brady, J. Brkljacic, J. Browse, C. Chapple, V. Colot, S. Cutler, J. Dangl, D. Ehrhardt, J. D. Friesner, W. B. Frommer, E. Grotewold, E. Meyerowitz, J. Nemhauser, M. Nordborg, C. Pikaard, J. Shanklin, C. Somerville, M. Stitt, K. U. Torii, J. Waese, D. Wagner, P. McCourt, 50 years of Arabidopsis research: highlights and future directions. New Phytol. 209, 921–944 (2016).
- S. Kagale, C. Koh, J. Nixon, V. Bollina, W. E. Clarke, R. Tuteja, C. Spillane, S. J. Robinson, M. G. Links, C. Clarke, E. E. Higgins, T. Huebert, A. G. Sharpe, I. A. P. Parkin, The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5, 3706 (2014).
- K. M. Dorn, J. D. Fankhauser, D. L. Wyse, M. D. Marks, A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Res. 22, 121–131 (2015).
- B. Chalhoub, F. Denoeud, S. Liu, I. A. P. Parkin, H. Tang, X. Wang, J. Chiquet, H. Belcram, C. Tong, B. Samans, M. Corréa, C. Da Silva, J. Just, C. Falentin, C. S. Koh, I. Le Clainche, M. Bernard, P. Bento, B. Noel, K. Labadie, A. Alberti, M. Charles, D. Arnaud, H. Guo, C. Daviaud, S. Alamery, K. Jabbari, M. Zhao, P. P. Edger, H. Chelaifa, D. Tack, G. Lassalle, I. Mestiri, N. Schnel, M.-C. Le Paslier, G. Fan, V. Renault, P. E. Bayer, A. A. Golicz, S. Manoli, T.-H. Lee, V. H. D. Thi, S. Chalabi, Q. Hu, C. Fan, R. Tollenaere, Y. Lu, C. Battail, J. Shen, C. H. D. Sidebottom, X. Wang, A. Canaguier, A. Chauveau, A. Bérard, G. Deniot, M. Guan, Z. Liu, F. Sun, Y. P. Lim, E. Lyons, C. D. Town, I. Bancroft, X. Wang, J. Meng, J. Ma, J. C. Pires, G. J. King, D. Brunel, R. Delourme, M. Renard, J.-M. Aury, K. L. Adams, J. Batley, R. J. Snowdon, J. Tost, D. Edwards, Y. Zhou, W. Hua, A. G. Sharpe, A. H. Paterson, C. Guan, P. Wincker, Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 345, 950–953 (2014).
- X. Gan, A. Hay, M. Kwantes, G. Haberer, A. Hallab, R. D. Ioio, H. Hofhuis, B. Pieper, M. Cartolano, U. Neumann, L. A. Nikolov, B. Song, M. Hajheidari, R. Briskine, E. Kougioumoutzi, D. Vlad, S. Broholm, J. Hein, K. Meksem, D. Lightfoot, K. K. Shimizu, R. Shimizu-Inatsugi, M. Imprialou, D. Kudrna, R. Wing, S. Sato, P. Huijser, D. Filatov, K. F. X. Mayer, R. Mott, M. Tsiantis, The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants. 2, 16167 (2016).
- C.-Y. Cheng, V. Krishnakumar, A. P. Chan, F. Thibaud-Nissen, S. Schobel, C. D. Town, Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017).
- M. Dassanayake, D.-H. Oh, J. S. Haas, A. Hernandez, H. Hong, S. Ali, D.-J. Yun, R. A. Bressan, J.-K. Zhu, H. J. Bohnert, J. M. Cheeseman, The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918 (2011).
- H.-J. Wu, Z. Zhang, J.-Y. Wang, D.-H. Oh, M. Dassanayake, B. Liu, Q. Huang, H.-X. Sun, R. Xia, Y. Wu, Y.-N. Wang, Z. Yang, Y. Liu, W. Zhang, H. Zhang, J. Chu, C. Yan, S. Fang, J. Zhang, Y. Wang, F. Zhang, G. Wang, S. Y. Lee, J. M. Cheeseman, B. Yang, B. Li, J. Min, L. Yang, J. Wang, C. Chu, S.-Y. Chen, H. J. Bohnert, J.-K. Zhu, X.-J. Wang, Q. Xie, Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U. S. A. 109, 12219–12224 (2012).
- R. Yang, D. E. Jarvis, H. Chen, M. A. Beilstein, J. Grimwood, J. Jenkins, S. Shu, S. Prochnik, M. Xin, C. Ma, J. Schmutz, R. A. Wing, T. Mitchell-Olds, K. S. Schumaker, X. Wang, The Reference Genome of the Halophytic Plant Eutrema salsugineum. Front. Plant Sci. 4, 46 (2013).
- R. V. Briskine, T. Paape, R. Shimizu-Inatsugi, T. Nishiyama, S. Akama, J. Sese, K. K. Shimizu, Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol. Ecol. Resour. 17, 1025–1036 (2017).
- E.-M. Willing, V. Rawat, T. Mandáková, F. Maumus, G. V. James, K. J. V. Nordström, C. Becker, N. Warthmann, C. Chica, B. Szarzynska, M. Zytnicki, M. C. Albani, C. Kiefer, S. Bergonzi, L. Castaings, J. L. Mateos, M. C. Berns, N. Bujdoso, T. Piofczyk, L. de Lorenzo, C. Barrero-Sicilia, I. Mateos, M. Piednoël, J. Hagmann, R. Chen-Min-Tao, R. Iglesias-Fernández, S. C. Schuster, C. Alonso-Blanco, F. Roudier, P. Carbonero, J. Paz-Ares, S. J. Davis, A. Pecinka, H. Quesneville, V. Colot, M. A. Lysak, D. Weigel, G. Coupland, K. Schneeberger, Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants. 1, 14023 (2015).
- A. Haudry, A. E. Platts, E. Vello, D. R. Hoen, M. Leclercq, R. J. Williamson, E. Forczek, Z. Joly-Lopez, J. G. Steffen, K. M. Hazzouri, K. Dewar, J. R. Stinchcombe, D. J. Schoen, X. Wang, J. Schmutz, C. D. Town, P. P. Edger, J. C. Pires, K. S. Schumaker, D. E. Jarvis, T. Mandáková, M. A. Lysak, E. van den Bergh, M. E. Schranz, P. M. Harrison, A. M. Moses, T. E. Bureau, S. I. Wright, M. Blanchette, An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45, 891–898 (2013).
- T. Slotte, K. M. Hazzouri, J. A. Ågren, D. Koenig, F. Maumus, Y.-L. Guo, K. Steige, A. E. Platts, J. S. Escobar, L. K. Newman, W. Wang, T. Mandáková, E. Vello, L. M. Smith, S. R. Henz, J. Steffen, S. Takuno, Y. Brandvain, G. Coop, P. Andolfatto, T. T. Hu, M. Blanchette, R. M. Clark, H. Quesneville, M. Nordborg, B. S. Gaut, M. A. Lysak, J. Jenkins, J. Grimwood, J. Chapman, S. Prochnik, S. Shu, D. Rokhsar, J. Schmutz, D. Weigel, S. I. Wright, The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835 (2013).
- Y. Sun, D.-H. Oh, L. Duan, P. Ramachandran, A. Ramirez, A. Bartlett, K.-N. Tran, G. Wang, M. Dassanayake, J. R. Dinneny, Divergence in the ABA gene regulatory network underlies differential growth control. Nat Plants (2022), doi:10.1038/s41477-022-01139-5.
- R. C. O’Malley, S.-S. C. Huang, L. Song, M. G. Lewsey, A. Bartlett, J. R. Nery, M. Galli, A. Gallavotti, J. R. Ecker, Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 165, 1280–1292 (2016).
- A. Bartlett, R. C. O’Malley, S.-S. C. Huang, M. Galli, J. R. Nery, A. Gallavotti, J. R. Ecker, Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12, 1659–1672 (2017).
- A. H. K. Roeder, P. T. Tarr, C. Tobin, X. Zhang, V. Chickarmane, A. Cunha, E. M. Meyerowitz, Computational morphodynamics of plants: integrating development over space and time. Nat. Rev. Mol. Cell Biol. 12, 265–273 (2011).
- L. M. York, E. A. Nord, J. P. Lynch, Integration of root phenes for soil resource acquisition. Front. Plant Sci. 4, 355 (2013).
- Y. Gao, J. P. Lynch, Reduced crown root number improves water acquisition under water deficit stress in maize (Zea mays L.). J. Exp. Bot. 67, 4545–4557 (2016).
- W. A. V. Beardall, G.-B. Stan, M. J. Dunlop, Deep Learning Concepts and Applications for Synthetic Biology. GEN Biotechnol. 1, 360–371 (2022).
- Ž. Avsec, V. Agarwal, D. Visentin, J. R. Ledsam, A. Grabska-Barwinska, K. R. Taylor, Y. Assael, J. Jumper, P. Kohli, D. R. Kelley, Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods. 18, 1196–1203 (2021).
- A. Shrikumar, P. Greenside, A. Kundaje, Learning Important Features Through Propagating Activation Differences. arXiv [cs.CV] (2017), (available at http://arxiv.org/abs/1704.02685).
- M. Exposito-Alonso, 500 Genomes Field Experiment Team, H. A. Burbano, O. Bossdorf, R. Nielsen, D. Weigel, Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature. 574, E16 (2019).
- J. A. N. Brophy, K. J. Magallon, L. Duan, V. Zhong, P. Ramachandran, K. Kniazev, J. R. Dinneny, Synthetic genetic circuits as a means of reprogramming plant roots. Science. 377, 747–751 (2022).
- T. P. Brutnell, L. Wang, K. Swartwood, A. Goldschmidt, D. Jackson, X.-G. Zhu, E. Kellogg, J. Van Eck, Setaria viridis: a model for C4 photosynthesis. Plant Cell. 22, 2537–2544 (2010).
- N. Wang, L. Ryan, N. Sardesai, E. Wu, B. Lenderts, K. Lowe, P. Che, A. Anand, A. Worden, D. van Dyk, P. Barone, S. Svitashev, T. Jones, W. Gordon-Kamm, Leaf transformation for efficient random integration and targeted genome modification in maize and sorghum. Nat Plants. 9, 255–270 (2023).
- T. Colombi, A. Chakrawal, A. M. Herrmann, Carbon supply-consumption balance in plant roots: effects of carbon use efficiency and root anatomical plasticity. New Phytol. 233, 1542–1547 (2022).
- J. A. Postma, A. Dathe, J. P. Lynch, The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 166, 590–602 (2014).
- T. LaRue, H. Lindner, A. Srinivas, M. Exposito-Alonso, G. Lobet, J. R. Dinneny, Uncovering natural variation in root system architecture and growth dynamics using a robotics-assisted phenomics platform, , doi:10.1101/2021.11.13.468476.
- R. Rellán-Álvarez, G. Lobet, H. Lindner, P.-L. Pradier, J. Sebastian, M.-C. Yee, Y. Geng, C. Trontin, T. LaRue, A. Schrager-Lavelle, C. H. Haney, R. Nieu, J. Maloof, J. P. Vogel, J. R. Dinneny, GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife. 4 (2015), doi:10.7554/eLife.07597.
- E. C. Morris, M. Griffiths, A. Golebiowska, S. Mairhofer, J. Burr-Hersey, T. Goh, D. von Wangenheim, B. Atkinson, C. J. Sturrock, J. P. Lynch, K. Vissenberg, K. Ritz, D. M. Wells, S. J. Mooney, M. J. Bennett, Shaping 3D Root System Architecture. Curr. Biol. 27, R919–R930 (2017).